DATA146
Project 1
Question 1
A Python package is a collection that contains multiple related modules and subpackages. Modules that are related to each other are usually put into the same package to reach a common goal. When a module from an external package is required in a program, that package can be imported and its modules can be put to use. A package is a directory of Python modules containing an additional __init__.py file, which distinguishes a package from a directory that happens to contain a bunch of Python scripts.
A Python library refers to the Standard Library, which is a large collection of modules and packages. A library comes with Python and is installed along with it, making its modules available to any Python code. One is able to use the modules in the library without having to download them from anywhere else.
In order to install a package and make the library of functions accessible to your local workspace, you have to first install the packages using the command pip install function_name.
pip install pandas
pip install numpy
Then you will have to import the packages using the command import package_name.
import pandas as pd
import numpy as np
If you want to call the functions inside that package, you can simply do so by using the dot(.) operator.
input = np.array(['cat’,'dog','fish'])
new_dataframe = pd.DataFrame(input, index=[1,2,3])
It is more convenient for you to use an alias such as pd and np because it allows you to call functions from the imported package using the short name, rather than having to type out the full name of the packages each time you want to call a function from it.
Question 2
A data frame is a two-dimensional, tabular data structure with labeled rows and columns. It is the most commonly used pandas object. Pandas is the library that is particularly useful for working with data frames and data analysis in general. After importing the pandas library, you need to read in a file. In order to read a file in its remote location within the file system of your operating system, you should use the pandas function read_csv( ).
import pandas as pd
data = pd.read_csv(‘gapminder.tsv’, sep = ‘\t’)
We need to specify the argument for the read_csv( ) function because we have to let it know which file to read and which type of file it is. First, we need to pass in the file. Then, it is important that when we are using the read_csv( ) function we need to know the type of the file. If the file we are importing is a tab-separated file, we have to add an additional argument following the file name. This is because by default the read_csv( ) function assumes that the file is a comma-separated file. We need to use the \t specification to inform python that our file is tab-separated.
In order to describe the data frame we just created, we can use the head() , info(), describe() functions.
data.head()
By using the head() function, we are able to see the top five rows of the new data frame. There are columns for country, continent, year, life expectancy, population, and GDP per capita.
data.info()
data.describe()
We can use data.info() to get a quick summary of the data. It shows you the names of the columns and the data types. We can also use data.describe() to see the descriptive statistics such as minimum, maximum, or mean of each column containing the numerical data.
To determine the number of rows and columns in the data frame, we can simply use the .shape command. By typing data.shape[0] and data.shape[1] in the console, we can see that there are 1704 rows and 7 columns (index included) respectively.
The alternate terminology for describing rows and columns is observations and variables respectively. Rows contain different information about an individual thing such as years, countries, and continents. Columns contain variables of the same sort of thing such as the year of 2002 and 2007.
Question 3
gapminder = pd.read_csv(‘gapminder.tsv’, sep = \t)
If we want to interrogate the year variable within the data frame, we can use the unique() function to check all the unique years in the data frame. The command for this would be data['year'].unique(). From the result we print out, we can see that the year variable does exhibit regular intervals of five years. If I were to add new outcomes to the raw data for updates, I would have to add the year of 2012 and 2017 according to the five year regular interval.
len(data['country'].unique())*2
To identify the number of new outcomes you would be adding to the data frame, you can first determine the number of countries in the data frame using the unique() function. By using the len() function, you are able to know the number of all the unique years. Last, since you are adding the data of two years to the data frame, we have to multiply the result by 2. And the answer would be 284.
Question 4
lowest = data[data['lifeExp']==data['lifeExp'].min()]
If you want to know which country at what point in time had the lowest life expectancy, we can use the command data[data['lifeExp']==data['lifeExp'].min()] and assign it to a new object “lowest” to see the result. Thus, by subsetting the gapminder data frame, we know that Rwanda in 1992 had the lowest life expectancy of 23.599 in the entire data. This is an unsurprising result because Rwanda was faced with a large-scale civil war between 1990 and 1994. The war arose as a consequence of the years-long dispute between the Hutu and Tutsi ethnicities, which eventually led to the infamous Rwanda genocide in 1994 with the death toll amounting to an astonishing 800,000. With mass killings plaguing Rwanda, it is reasonable that its life expectancy in 1992 dropped to a global minimum of 23.599.
Question 5
gdp = data['pop']*data['gdpPercap']
data.insert(6,'gdp',gdp, True)
data.sort_values(by=['gdp'], inplace=True, ascending=False)
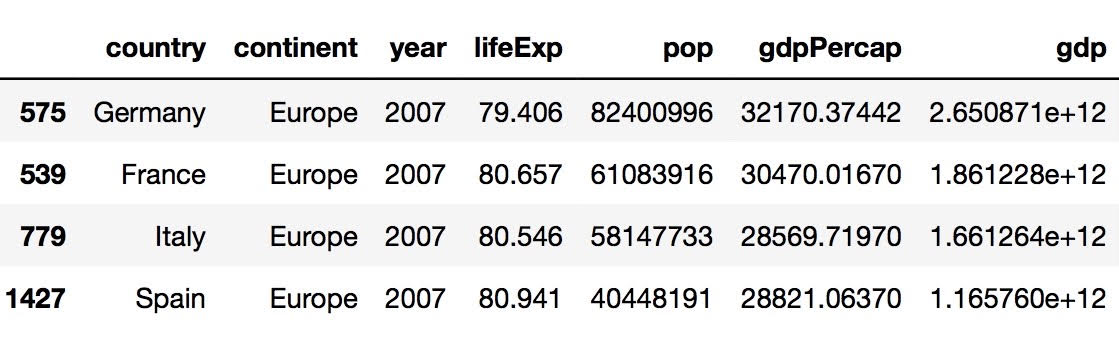
gdp_europe = data[data["country"].isin(["Germany","France","Spain","Italy"])&data['year'].isin(['2007'])]
To multiply the variable pop by the variable gdpPercap, you can simply use the command data['pop']*data['gdpPercap'] and assign the result to a new object called “gdp.” Since we want to add the new data of gdp to the existing data frame, I used the insert() function by using the command data.insert(6,'gdp',gdp, True). The argument 6 here represents the index of the new column to be added to the data frame. To sort the value of gdp from highest to lowest, I used the sort_values() function and passed in the necessary arguments. Since by default, the sort_values() function would assume that we want to sort by an ascending order, we have to pass in the ascending=False argument for it to sort from highest to lowest. To identify the gdp for Germany, France, Spain, and Italy in the year of 2007, I used the isin() function twice so that I was able to select the rows on the basis of country as well as year. Finally, I assigned the result to a new data frame called gdp_europe.
gdp_growth = data[data["country"].isin(["Germany","France","Spain","Italy"])&data['year'].isin(['2007','2002'])]
If we want to look at the most significant total gross domestic product increase during the previous 5-year period, we can simply do so by adding the year of 2002 to the argument for gdp_europe and assigning it to a new object called gdp_growth. From the result, we can tell that Germany had the greatest increase in GDP from 2002 to 2007.
Question 6
& stands for the AND operator, which returns True if both the operands are true. It is most commonly used with conditional statements. For example, if we want to select the observations for France in 2002 in the data frame, we can use the command below.
data.loc[(data['country'] == 'France') & (data['year'] == 2002)]
| is the inclusive OR operator, which returns True when either one of the operands is true. It is also a very common use for conditional statements. If we want to select the observations from either France or Spain, we can use the command below.
data.loc[(data['country'] == 'France') | (data['country'] == 'Spain')]
^ is the exclusive XOR operator, which returns True when only one of the two operands is true. One pitfall with using the ^ operator is that you would not know if only one or both of the statements are true. For example, the ^ operator returns False when ('dog'=='dog') ^ ('cat'=='cat').
The == logical operator is one of the comparison operators which is used to compare values. It returns True or False in response to the condition given. The == operator is checking if the two operands are equal to each other. The == operator can be used to compare numbers, strings, or functions. The == operator is most commonly used in Boolean statements which return True or False. The function of a single = sign is different from a double == sign because the former is an assignment operation. Below is an example of how to use the == operator.
x= 'cat'
y= 'dog'
x==y, returns False
Question 7
.loc is used in data frames and is used to access observations by labels or a boolean array. .loc takes index labels and returns a series or a data frame if the index label exists in the caller data frame. It returns a data frame or a series depending on the parameters. If you pass in a list as the parameter, the data type of the returned value would be a data frame. If you simply pass in the index label, then the output would be of a series type.
The .iloc attribute for data frames is used for integer-location based indexing or selection by position, meaning to select data by numbers. There are two required arguments for .iloc— a row selector and a column selector. .iloc returns a series when one row is selected, and a data frame when multiple rows are selected. Usually, .loc is more widely used than .iloc.
consecutive_rows = data.iloc[0:10] #rows
To extract a series of consecutive observations from a data frame, you can use the .iloc attribute. For example, to select the first 10 consecutive rows of data, use the command data.iloc[0:10].
consecutive_columns = data.iloc[:,0:3] #columns
To extract a series of consecutive columns, you can also use the .iloc attribute to achieve this. For example, to select the first three rows, use the command data.iloc[:,0:3]. Remember to pass “:” in to indicate that we are referring to the columns.
Question 8
API is short for “Application Programming Interface,” which is a set of rules that is shared by a particular service. These rules determine in which format or with which command set your application can access the service, as well as what data this service can return in the response. APIs are most commonly used to retrieve data. It acts as a layer between your application and external service.
To construct a request to a remote server in order to pull data, we have to first import the requests library and then assign the url for the desired data.
import requests
url= #the url containing the desired data
Then we have to create a new folder that will contain the downloaded data later.
# Use the os library for this
import os
#build a folder called 'data'
data_folder = 'data'
if not os.path.exists(data_folder):
os.makedirs(data_folder)
# Now construct the file name
file_name_short = 'ctp_' + str(dt.now(tz = pytz.utc)).replace(' ', '_') + '.csv'
file_name = os.path.join(data_folder, file_name_short)
After finishing the above steps, we are now able to retrieve the content from the url and open our output file for writing (w) in binary mode (b). Then we are all set to import the data we just downloaded as a pandas data frame.
r = requests.get(url)
with open(file_name, 'wb') as f:
f.write(r.content)
import pandas as pd
df = pd.read_csv(file_name)
Question 9
The purpose of the apply() function from the pandas library is to apply a function along an axis of a data frame. It takes a function as an input and applies this function to an entire data frame. One thing to note is that you have to specify the axis that you want the function to act on (0 for columns; and 1 for rows). The apply() function can also be used with anonymous functions or lambda functions. Using the apply() function to various class objects is an alternative to writing loops which have to iterate over each column. apply() could be a preferred approach because it offers convenience. It accepts any user defined function that applies a transformation to a data frame or a series. Also, it can be applied both row-wise and column-wise.
Question 10
An alternative approach to filtering the number of columns in a data frame is to use the .filter() function to the data frame. The filter() function can subset the rows or columns according to the specified index labels. In order to subset the desired data and return a new data frame, you have to specify the labels of the columns. Another approach to achieve this is to use Boolean expressions. For example, we can filter the gapminder data frame based on the year’s value of 2002. This conditional statement results in a boolean variable that has True when the value of year equals 2002. And then use the command data[data['year']==2002] and assign it to a new data frame. This will also successfully subset the data frame based on the year of 2002.
data.filter(items=['year','continent'])
2002data = data[data['year']==2002]